Software
Here are some software I have created and maintained.
causarray 
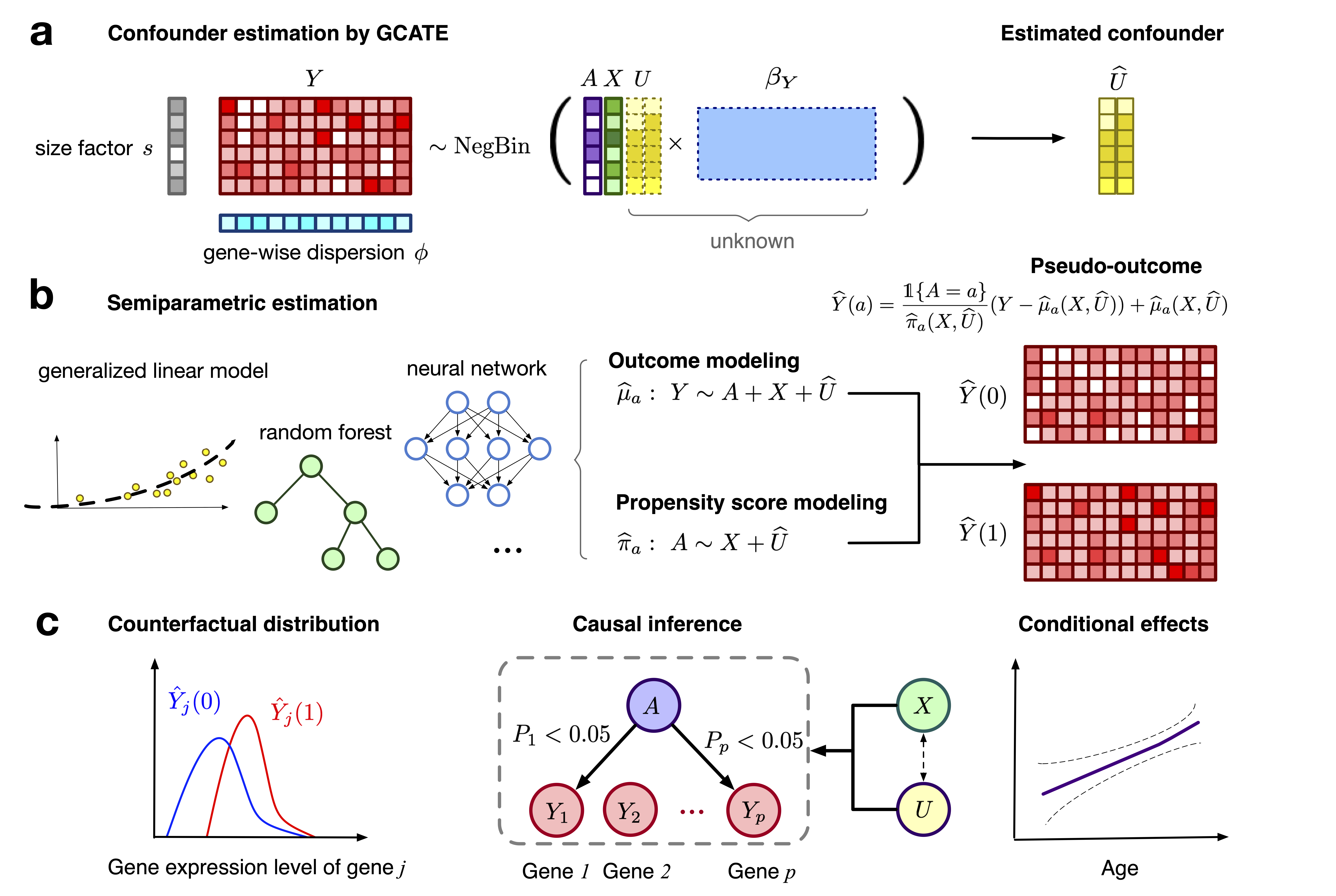
causarray is a Python package that applies causal inference methods to genomic studies. It is designed to identify genes that are causally affected by specific conditions, such as Alzheimer's disease (AD) and Autsim spectrum disorders and neurodevelopmental delay (ASD/ND), by controlling for both observed and unmeasured confounders in gene expression data.
- Confounder Adjustment: causarray utilizes a modified GCATE method to estimate unmeasured confounders adjusts for both observed and unmeasured confounders in gene expression data. This method enhances causal inference by directly estimating confounding effects without relying solely on negative control genes, which may not always be available or accurately identified.
- Doubly-Robust Counterfactual Inference: Utilizes the doubly-robust semiparametric framework for counterfactual imputation and inference. By modeling both the treatment assignment mechanism and the outcome, causarray remains consistent if either model is correctly specified, increasing the reliability of causal effect estimates in genomic studies.
- Causally Affected Expression Analysis: causarray focuses on identifying genes that are causally affected by specific conditions, such as diseases or treatments. This allows for unbiased differential expression analysis, improving the detection of true causal effects and providing deeper insights into the biological processes underlying conditions like Alzheimer's disease.
VITAE 
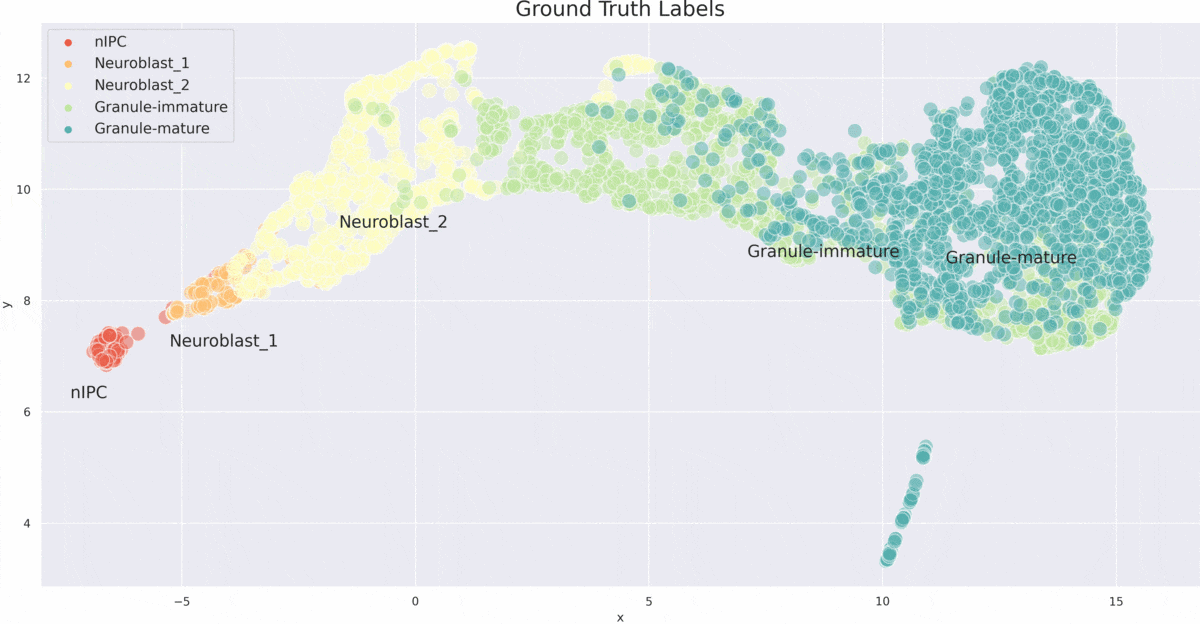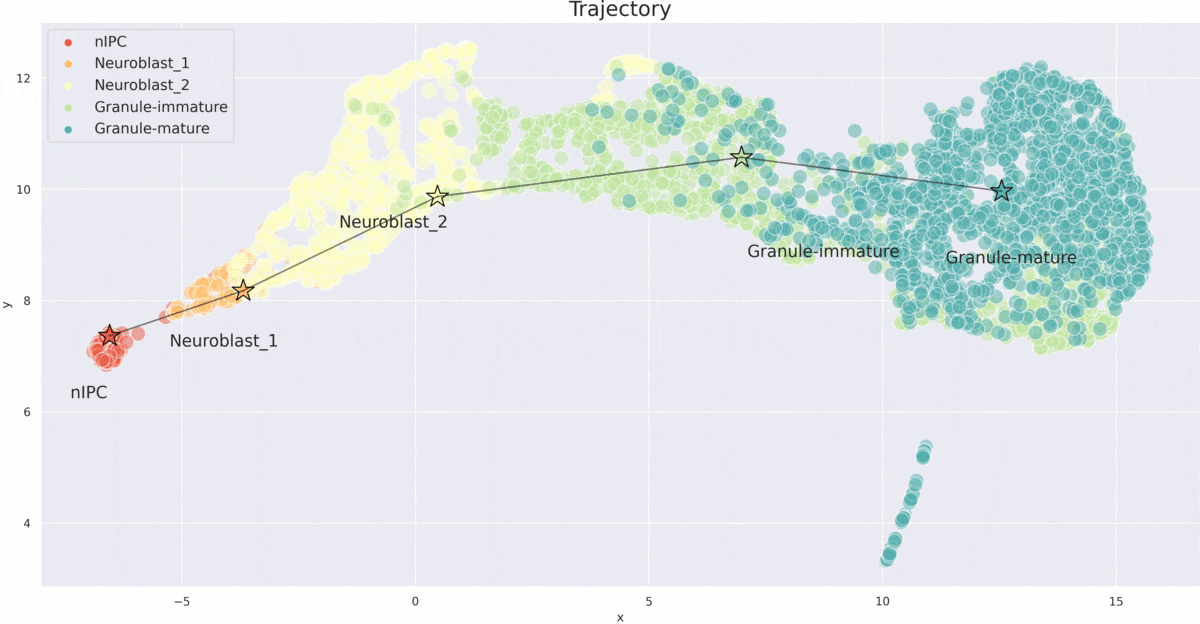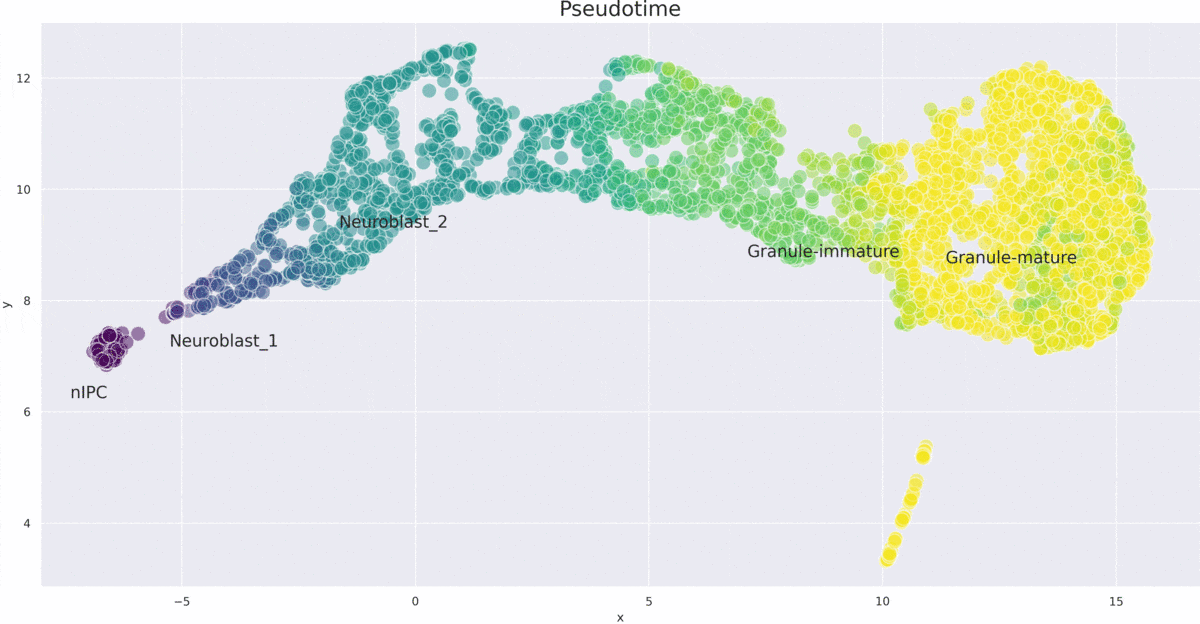
VITAE (Variational Inference for for Trajectory Analysis by AutoEncoder) is a method for inferring developmental trajectories from single-cell RNA-seq data. It integrates and aligns single-cell data from different modalities such as chromatin accessibility and gene expression.
- Trajectory Inference: Accurately infers developmental trajectories and pseudotime in various datasets.
- Data Integration: Better handles the integration of multiple single-cell datasets, adjusting for batch effects and other confounders.
- Accelerated Gaussian Version: An accelerated version that approximates distributions with Gaussian assumptions for computational efficiency.
- Differential Gene Expression Analysis: Effective in identifying differentially expressed genes along inferred trajectories.
scVAEIT 
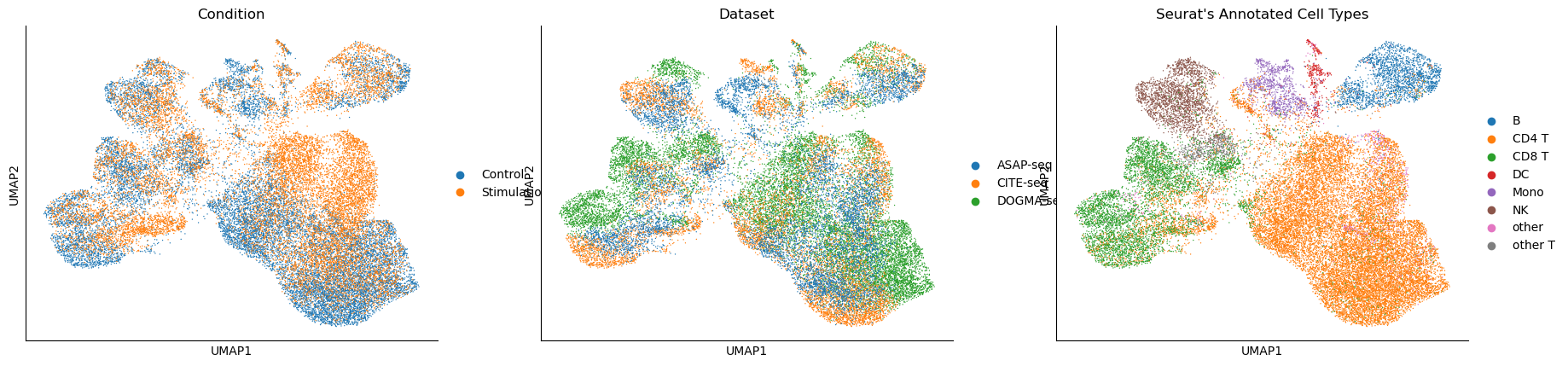
scVAEIT (single-cell Variational AutoEncoder for integration and transfer learning) is a Python module that utilizes a variational autoencoder (VAE) for single-cell mosaic integration and transfer learning. It aims to integrate and impute single-cell data from different modalities, such as gene expression, protein abundance, and chromatin accessibility.
- Multimodal Data Integration: Integrates single-cell data from multiple modalities, such as scRNA-seq, scATAC-seq, and CITE-seq, when the observations may not share the same set of features.
- Imputation: Imputes missing values in single-cell datasets by leveraging information from other modalities.
- Transfer Learning and Cross-modality Translation: Enables transfer learning by training on a reference dataset and readily transferring the learned knowledge to new sources for cross-modality translation and imputation.
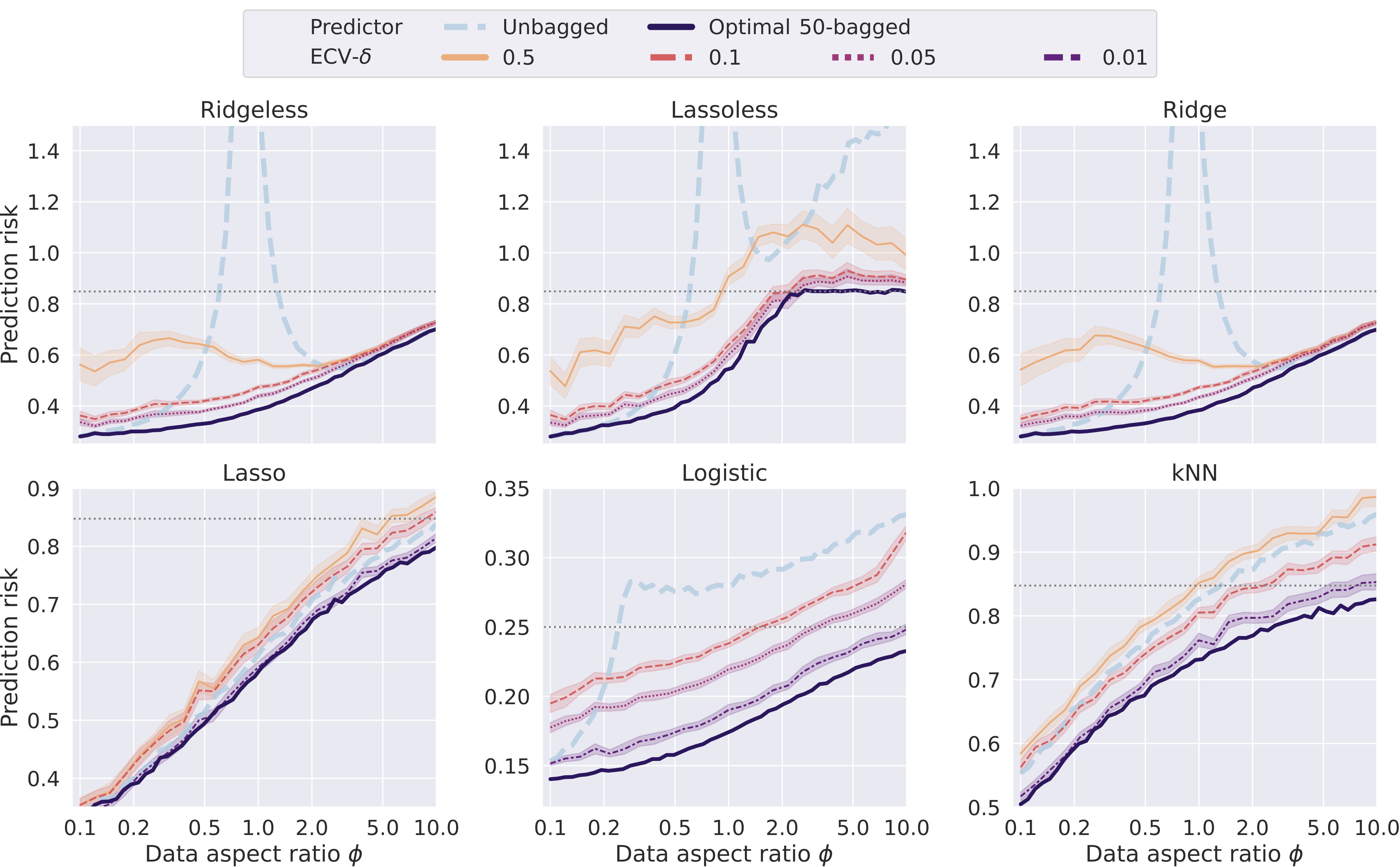
sklearn_ensemble_cv 
sklearn_ensemble_cv is a Python module for performing accurate and efficient ensemble cross-validation methods from various projects.
- Flexibility: The module builds on scikit-learn/sklearn to provide the most flexibility on various base predictors.
- Risk Estimation and Hyperparameter Tuning for Ensemble Learning: The module includes functions for creating ensembles of models, training the ensembles using cross-validation, and making predictions with the ensembles.
- Evaluation Utilities: The module also includes utilities for evaluating the performance of the ensembles and the individual models that make up the ensembles.